Calculates rate of oxygen uptake or production from respirometry data. A rate
can be determined over the whole dataset, or on subsets of the data using the
from and to inputs to specify data regions in terms of oxygen or time
units or row numbers of the input data. Multiple rates can be extracted
from the same dataset by using these inputs to enter vectors of paired values
in the appropriate metric. See Examples.
Arguments
- x
object of class
inspectordata.frame. This is the timeseries of paired values of oxygen against time from which to calculate rates.- from
numeric value or vector. Defaults to
NULL. The start of the region(s) over which you want to calculate the rate in the units specified inby. If a vector, each value must have a paired value into.- to
numeric value or vector. Defaults to
NULL. The end of the region(s) over which you want to calculate the rate in the units specified inby. If a vector, each value must have a paired value infrom.- by
string.
"time","row", or"oxygen". Defaults to"time". This is the method used to subset the data region betweenfromandto.- plot
logical. Defaults to
TRUE. Plot the results.- ...
Allows additional plotting controls to be passed, such as
pos,panel, andquiet = TRUE.
Value
Output is a list object of class calc_rate containing input
parameters and data, various summary data, metadata, linear models, and the
primary output of interest $rate, which can be background adjusted in
adjust_rate or converted to units in convert_rate.
Details
The function calculates rates by fitting a linear model of oxygen against
time, with the slope of this regression being the rate. There are no units
involved in calc_rate. This is a deliberate decision. The units of oxygen
concentration and time will be specified later in convert_rate() when
rates are converted to specific output units.
For continuous data recordings, it is recommended a data.frame containing
the data be prepared via inspect(), and entered as the x input. For
data not prepared like this, x can be a 2-column data.frame containing
numeric values of time (col 1) and oxygen (col 2). If multiple columns are
found in either an inspect or data frame input, only the first two columns
are used.
Specifying regions
For calculating rates over specific regions of the data, the from and to
inputs in the by units of "time" (the default), "oxygen", or "row".
The from and to inputs do not need to be precise; the function will use
the closest values found.
Multiple regions can be examined within the same dataset by entering from
and to as vectors of paired values to specify different regions. In this
case, $rate in the output will be a vector of multiple rates with each
result corresponding to the position of the paired from and to inputs. If
from and to are NULL (the default), the rate is determined over the
entire dataset.
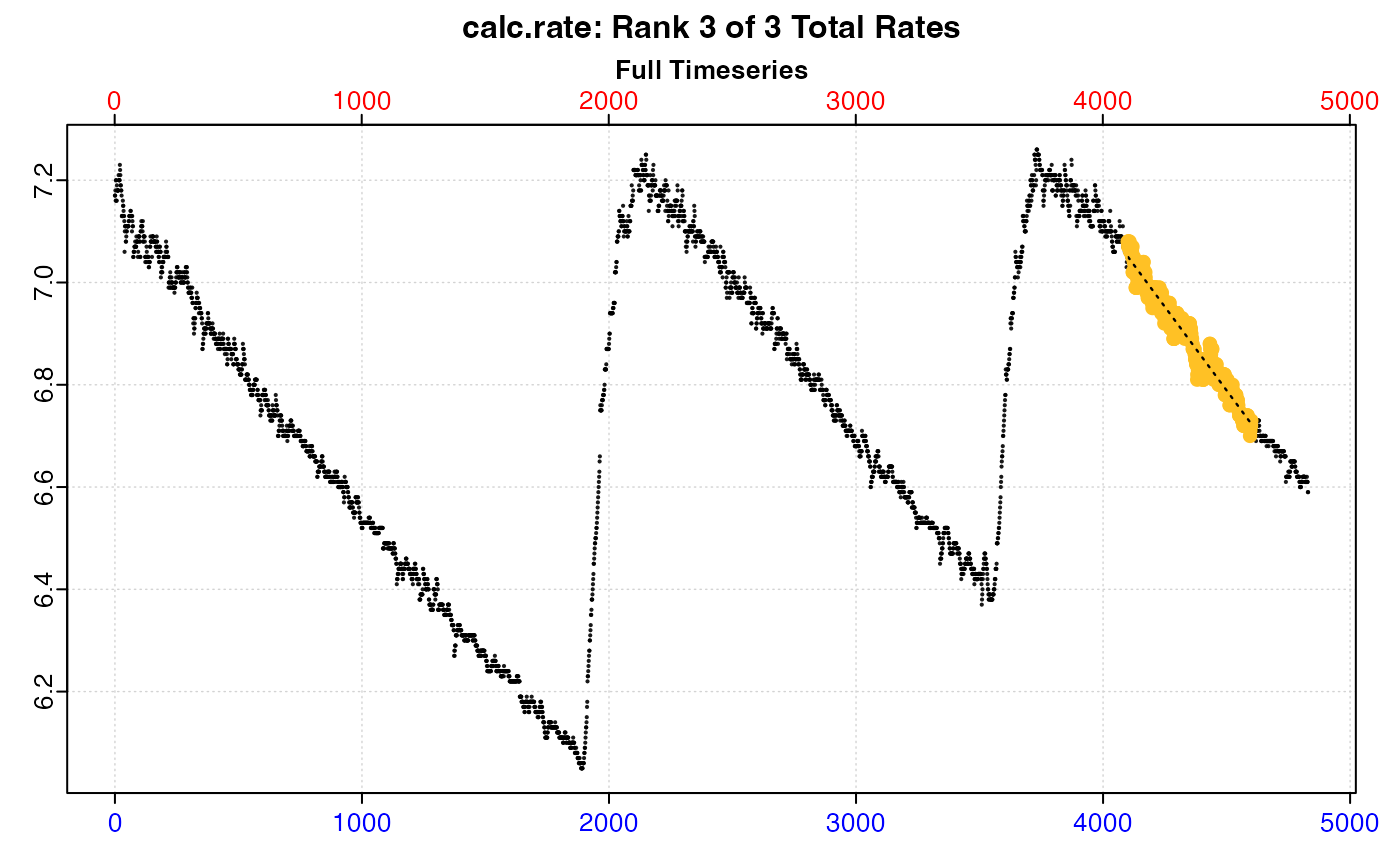
Plot
A plot is produced (provided plot = TRUE) showing the original data
timeseries of oxygen against time (bottom blue axis) and row index (top red
axis), with the region specified via the from and to inputs highlighted.
Second panel is a close-up of the rate region with linear model coefficients.
Third and fourth panels are summary plots of fit and residuals.
Additional plotting options
If multiple rates have been calculated, by default the first (pos = 1) is
plotted. Others can be plotted by changing the pos input either in the main
function call, or by plotting the output, e.g. plot(object, pos = 2). In
addition, each sub-panel can be examined individually by using the panel
input, e.g. plot(object, panel = 2).
Console output messages can be suppressed using quiet = TRUE. If axis
labels (particularly y-axis) are difficult to read, las = 2 can be passed
to make axis labels horizontal, and oma (outer margins, default oma = c(0.4, 1, 1.5, 0.4)), and mai (inner margins, default mai = c(0.3, 0.15, 0.35, 0.15)) used to adjust plot margins.
S3 Generic Functions
Saved output objects can be used in the generic S3 functions print(),
summary(), and mean().
print(): prints a single result, by default the first rate. Others can be printed by passing theposinput. e.g.print(x, pos = 2)summary(): prints summary table of all results and metadata, or those specified by theposinput. e.g.summary(x, pos = 1:5). The summary can be exported as a separate dataframe by passingexport = TRUE.mean(): calculates the mean of all rates, or those specified by theposinput. e.g.mean(x, pos = 1:5)The mean can be exported as a separate value by passingexport = TRUE.
More
For additional help, documentation, vignettes, and more visit the respR
website at https://januarharianto.github.io/respR/
Examples
# Subset by 'time' (the default)
inspect(sardine.rd, time = 1, oxygen = 2, plot = FALSE) %>%
calc_rate(from = 200, to = 1800)
#> inspect: No issues detected while inspecting data frame.
#>
#> # print.inspect # -----------------------
#> Time Oxygen
#> numeric pass pass
#> Inf/-Inf pass pass
#> NA/NaN pass pass
#> sequential pass -
#> duplicated pass -
#> evenly-spaced pass -
#>
#> -----------------------------------------
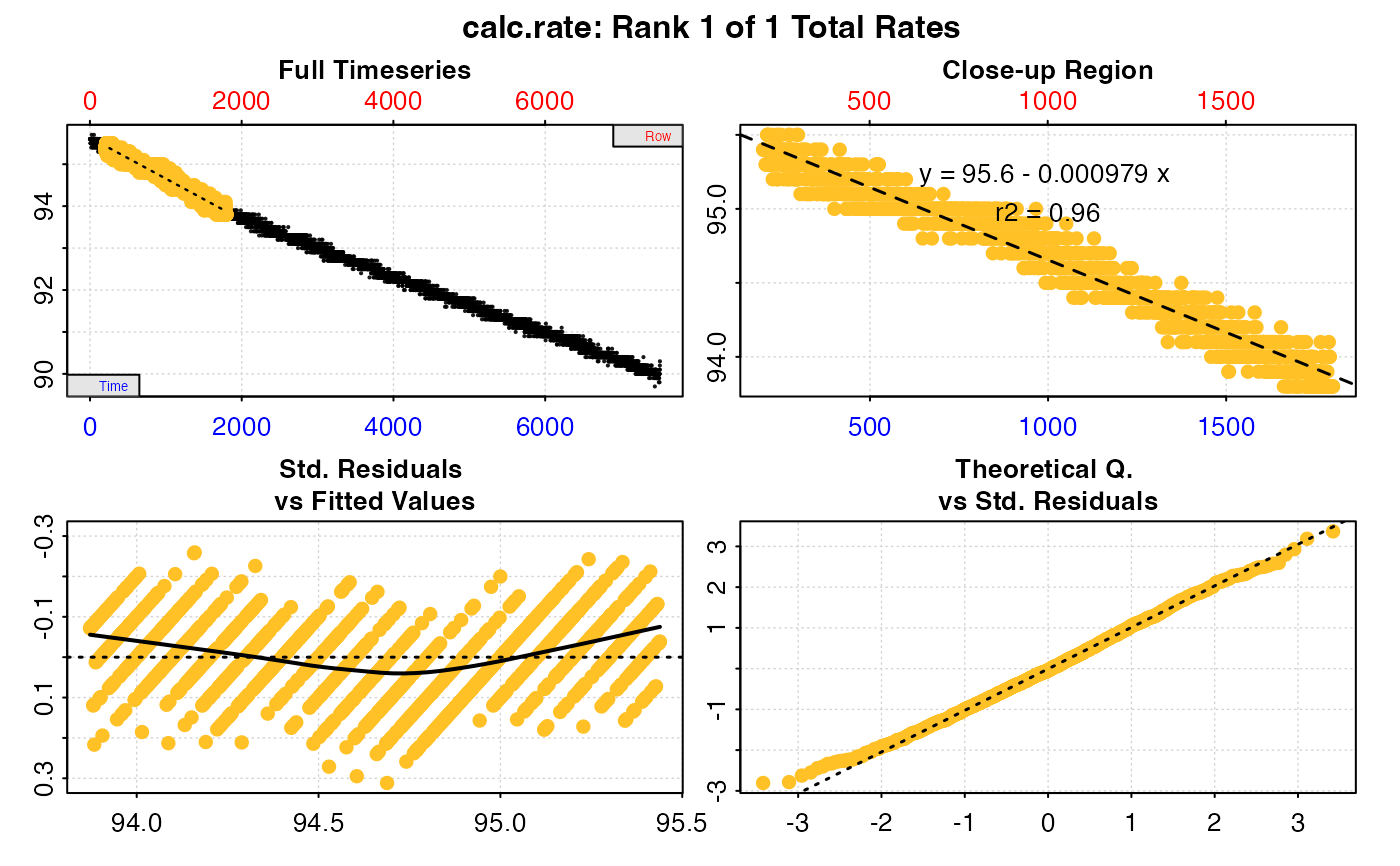 #>
#> # print.calc_rate # ---------------------
#> Rank 1 of 1 rates:
#> Rate: -0.0009793698
#>
#> To see full results use summary().
#> -----------------------------------------
# Subset by oxygen
inspect(sardine.rd, time = 1, oxygen = 2, plot = FALSE) %>%
calc_rate(94, 91, by = "oxygen")
#> inspect: No issues detected while inspecting data frame.
#>
#> # print.inspect # -----------------------
#> Time Oxygen
#> numeric pass pass
#> Inf/-Inf pass pass
#> NA/NaN pass pass
#> sequential pass -
#> duplicated pass -
#> evenly-spaced pass -
#>
#> -----------------------------------------
#>
#> # print.calc_rate # ---------------------
#> Rank 1 of 1 rates:
#> Rate: -0.0009793698
#>
#> To see full results use summary().
#> -----------------------------------------
# Subset by oxygen
inspect(sardine.rd, time = 1, oxygen = 2, plot = FALSE) %>%
calc_rate(94, 91, by = "oxygen")
#> inspect: No issues detected while inspecting data frame.
#>
#> # print.inspect # -----------------------
#> Time Oxygen
#> numeric pass pass
#> Inf/-Inf pass pass
#> NA/NaN pass pass
#> sequential pass -
#> duplicated pass -
#> evenly-spaced pass -
#>
#> -----------------------------------------
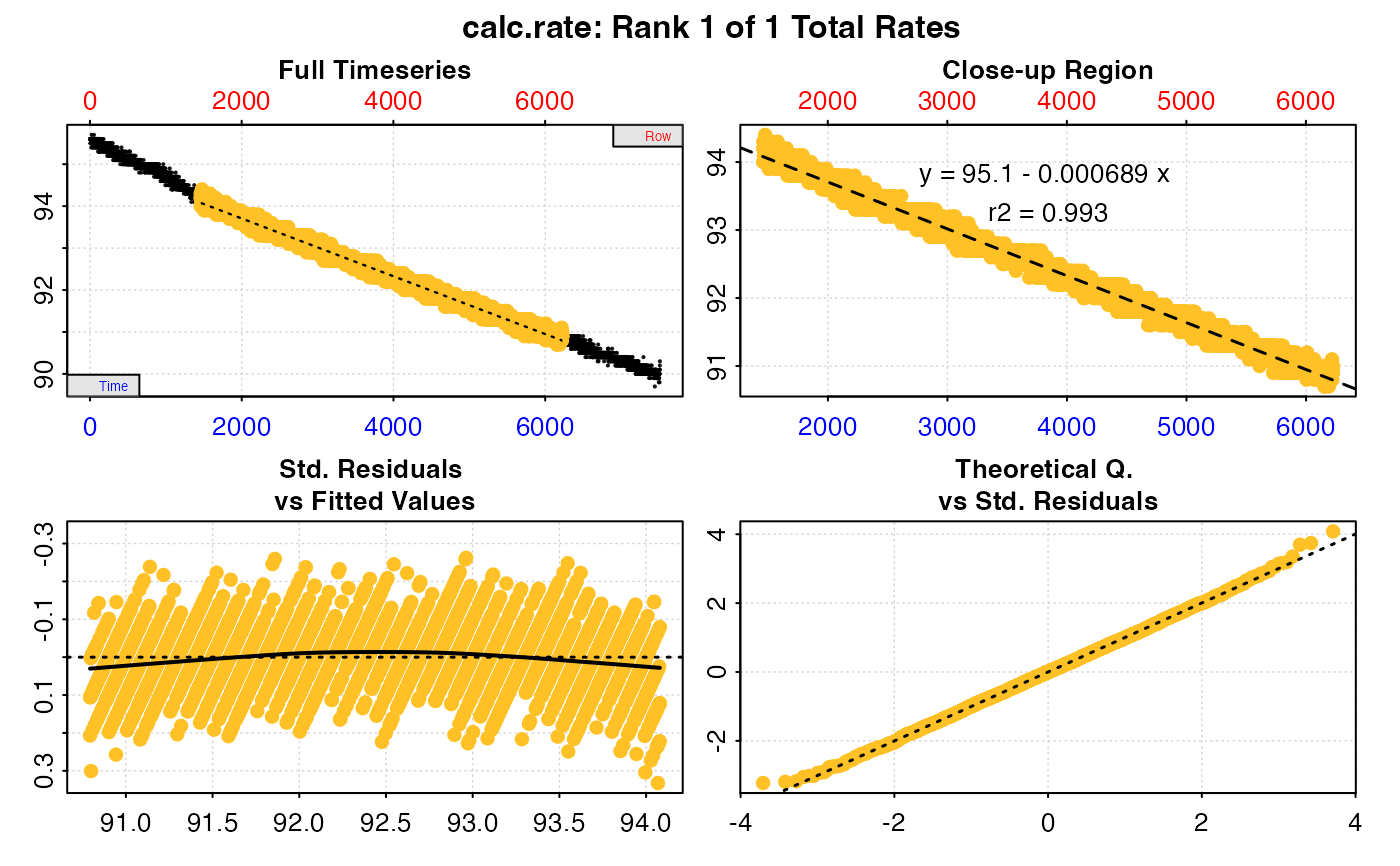 #>
#> # print.calc_rate # ---------------------
#> Rank 1 of 1 rates:
#> Rate: -0.0006892677
#>
#> To see full results use summary().
#> -----------------------------------------
# Subset by row
inspect(sardine.rd, time = 1, oxygen = 2, plot = FALSE) %>%
calc_rate(1000, 2000, by = "row")
#> inspect: No issues detected while inspecting data frame.
#>
#> # print.inspect # -----------------------
#> Time Oxygen
#> numeric pass pass
#> Inf/-Inf pass pass
#> NA/NaN pass pass
#> sequential pass -
#> duplicated pass -
#> evenly-spaced pass -
#>
#> -----------------------------------------
#>
#> # print.calc_rate # ---------------------
#> Rank 1 of 1 rates:
#> Rate: -0.0006892677
#>
#> To see full results use summary().
#> -----------------------------------------
# Subset by row
inspect(sardine.rd, time = 1, oxygen = 2, plot = FALSE) %>%
calc_rate(1000, 2000, by = "row")
#> inspect: No issues detected while inspecting data frame.
#>
#> # print.inspect # -----------------------
#> Time Oxygen
#> numeric pass pass
#> Inf/-Inf pass pass
#> NA/NaN pass pass
#> sequential pass -
#> duplicated pass -
#> evenly-spaced pass -
#>
#> -----------------------------------------
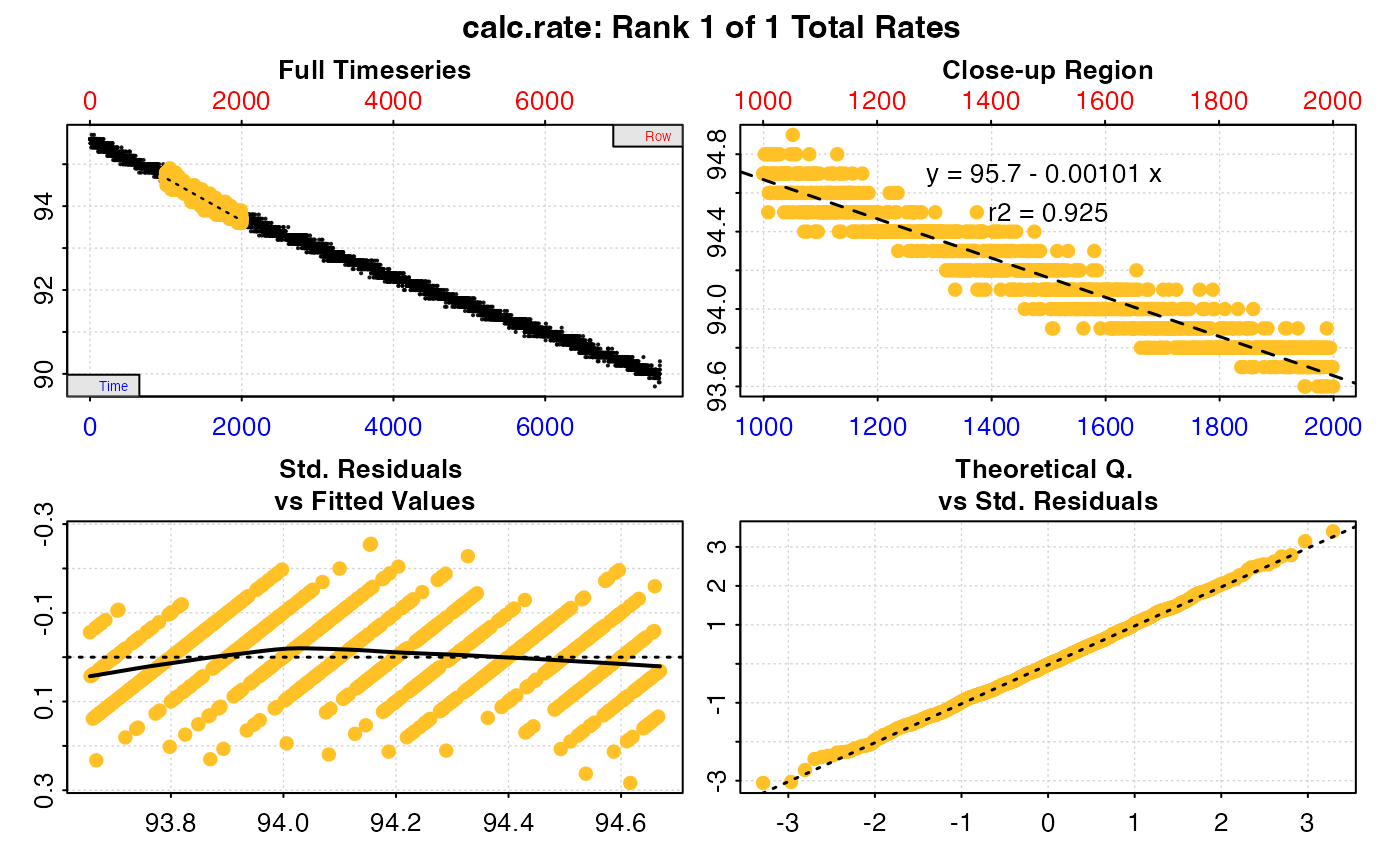 #>
#> # print.calc_rate # ---------------------
#> Rank 1 of 1 rates:
#> Rate: -0.001012827
#>
#> To see full results use summary().
#> -----------------------------------------
# Use a data frame input, and calculate rate from multiple regions by
# using a vector in the 'from' and 'to' inputs
x <- calc_rate(intermittent.rd,
from = c(200,2300,4100),
to = c(1800,3200,4600),
by = 'time',
plot = FALSE)
# Print and summary of results
print(x)
#>
#> # print.calc_rate # ---------------------
#> Rank 1 of 3 rates:
#> Rate: -0.0005734109
#>
#> To see other results use 'pos' input.
#> To see full results use summary().
#> -----------------------------------------
summary(x)
#>
#> # summary.calc_rate # -------------------
#> Summary of all rate results:
#>
#> rep rank intercept_b0 slope_b1 rsq row endrow time endtime oxy endoxy rate.2pt rate
#> 1: NA 1 7.127202 -0.0005734109 0.995 201 1801 200 1800 7.05 6.11 -0.0005875 -0.0005734109
#> 2: NA 2 8.528274 -0.0006097325 0.990 2301 3201 2300 3200 7.12 6.58 -0.0006000 -0.0006097325
#> 3: NA 3 9.731918 -0.0006539752 0.967 4101 4601 4100 4600 7.08 6.73 -0.0007000 -0.0006539752
#> -----------------------------------------
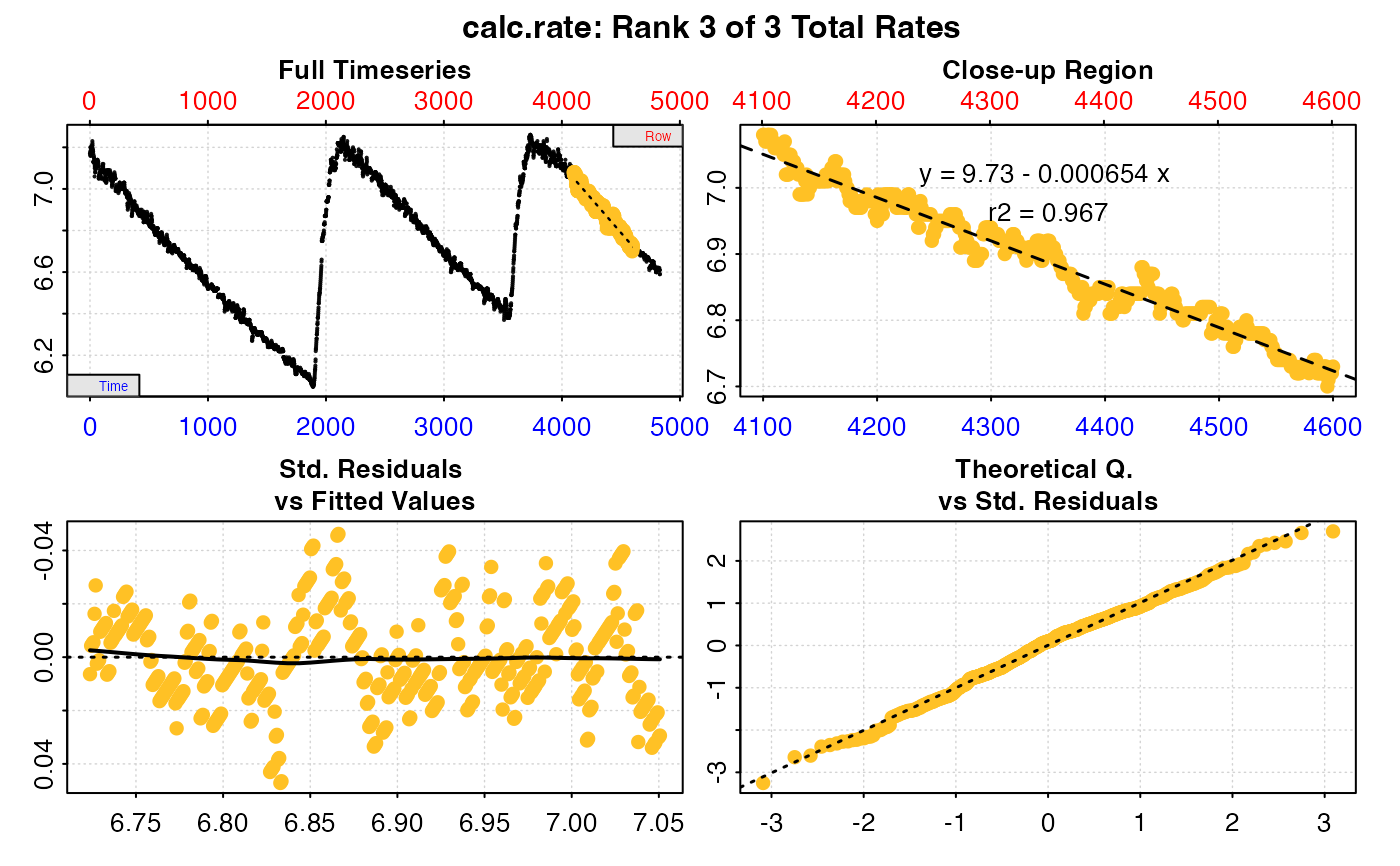
# Plot the third of these results
plot(x, pos = 3)
#>
#> # plot.calc_rate # ----------------------
#> plot.calc_rate: Plotting rate from position 3 of 3 ...
#>
#> # print.calc_rate # ---------------------
#> Rank 1 of 1 rates:
#> Rate: -0.001012827
#>
#> To see full results use summary().
#> -----------------------------------------
# Use a data frame input, and calculate rate from multiple regions by
# using a vector in the 'from' and 'to' inputs
x <- calc_rate(intermittent.rd,
from = c(200,2300,4100),
to = c(1800,3200,4600),
by = 'time',
plot = FALSE)
# Print and summary of results
print(x)
#>
#> # print.calc_rate # ---------------------
#> Rank 1 of 3 rates:
#> Rate: -0.0005734109
#>
#> To see other results use 'pos' input.
#> To see full results use summary().
#> -----------------------------------------
summary(x)
#>
#> # summary.calc_rate # -------------------
#> Summary of all rate results:
#>
#> rep rank intercept_b0 slope_b1 rsq row endrow time endtime oxy endoxy rate.2pt rate
#> 1: NA 1 7.127202 -0.0005734109 0.995 201 1801 200 1800 7.05 6.11 -0.0005875 -0.0005734109
#> 2: NA 2 8.528274 -0.0006097325 0.990 2301 3201 2300 3200 7.12 6.58 -0.0006000 -0.0006097325
#> 3: NA 3 9.731918 -0.0006539752 0.967 4101 4601 4100 4600 7.08 6.73 -0.0007000 -0.0006539752
#> -----------------------------------------
# Plot the third of these results
plot(x, pos = 3)
#>
#> # plot.calc_rate # ----------------------
#> plot.calc_rate: Plotting rate from position 3 of 3 ...
 #> -----------------------------------------
# Plot only the timeseries plot and hide the legend
plot(x, pos = 3, panel = 1, legend = FALSE)
#>
#> # plot.calc_rate # ----------------------
#> plot.calc_rate: Plotting rate from position 3 of 3 ...
#> -----------------------------------------
# Plot only the timeseries plot and hide the legend
plot(x, pos = 3, panel = 1, legend = FALSE)
#>
#> # plot.calc_rate # ----------------------
#> plot.calc_rate: Plotting rate from position 3 of 3 ...
 #> -----------------------------------------
#> -----------------------------------------
