
calc_rate.int: Manual extraction of rates in intermittent-flow respirometry
Source:vignettes/calc_rate.int.Rmd
calc_rate.int.RmdIntroduction
calc_rate.int() is a function for manually determining
rates across user-defined ranges of row or time in each replicate in
intermittent-flow respirometry data. This page contains descriptions and
simple examples of the functionality. See also the help file at
help("calc_rate.int"), see
vignette("intermittent_short") for how it can be used to
analyse a relatively brief intermittent-flow respirometry experiment,
and vignette("intermittent_long") for an example of
analysing a longer experiment.
Overview
How calc_rate.int works is fairly straightforward. The
function uses the starts locations to subset each replicate
from the data in x. It extracts a rate using the
wait and measure inputs in the by
metric.
A calc_rate object is saved for each replicate in the
output in $results, and the output $summary
table contains the results from all replicates in order with the
$rep column indicating the replicate number. This output
object can be passed to subsequent functions such as
adjust_rate() and convert_rate() for further
processing.
Inputs
There are two required inputs:
x: The time~oxygen data from a single intermittent-flow experiment as either adata.frameor aninspectobject-
starts: The location of the start of each replicate. This can be either:- A numeric vector of row numbers or times indicating each replicate start location.
- A single numeric value representing a regular row or time interval
starting from the first row or time value in the data. This option
should only be used when replicates cycle at regular intervals. If the
first replicate does not start at row 1 of the data in
xit should be subset so that it does. Seesubset_data()and examples below for how to do this.
Other inputs
wait: Rows or time period to exclude at the start of each replicate.measure: Rows or time period over which to calculate rate in each replicate. Applied directly afterwaitphase.by: Method by whichstarts,waitandmeasureare applied. Either"row"or"time".plot: Controls if a plot is produced. See Plot section.
Irregularly spaced replicates
Data
The urchin dataset used for examples below is the
included intermittent.rd example data with the time values
changed to minutes to better demonstrate time region selection.
Experimental data such as volume, weight, and row locations of
replicates, flushes etc. can be found in the data help file:
?intermittent.rd.
urchin <- intermittent.rd
urchin[[1]] <- round(urchin[[1]] / 60, 2) # change time values to minutes and round themThis is what the whole dataset look like. There are three replicates, and note they are of different duration.
urchin <- inspect(urchin)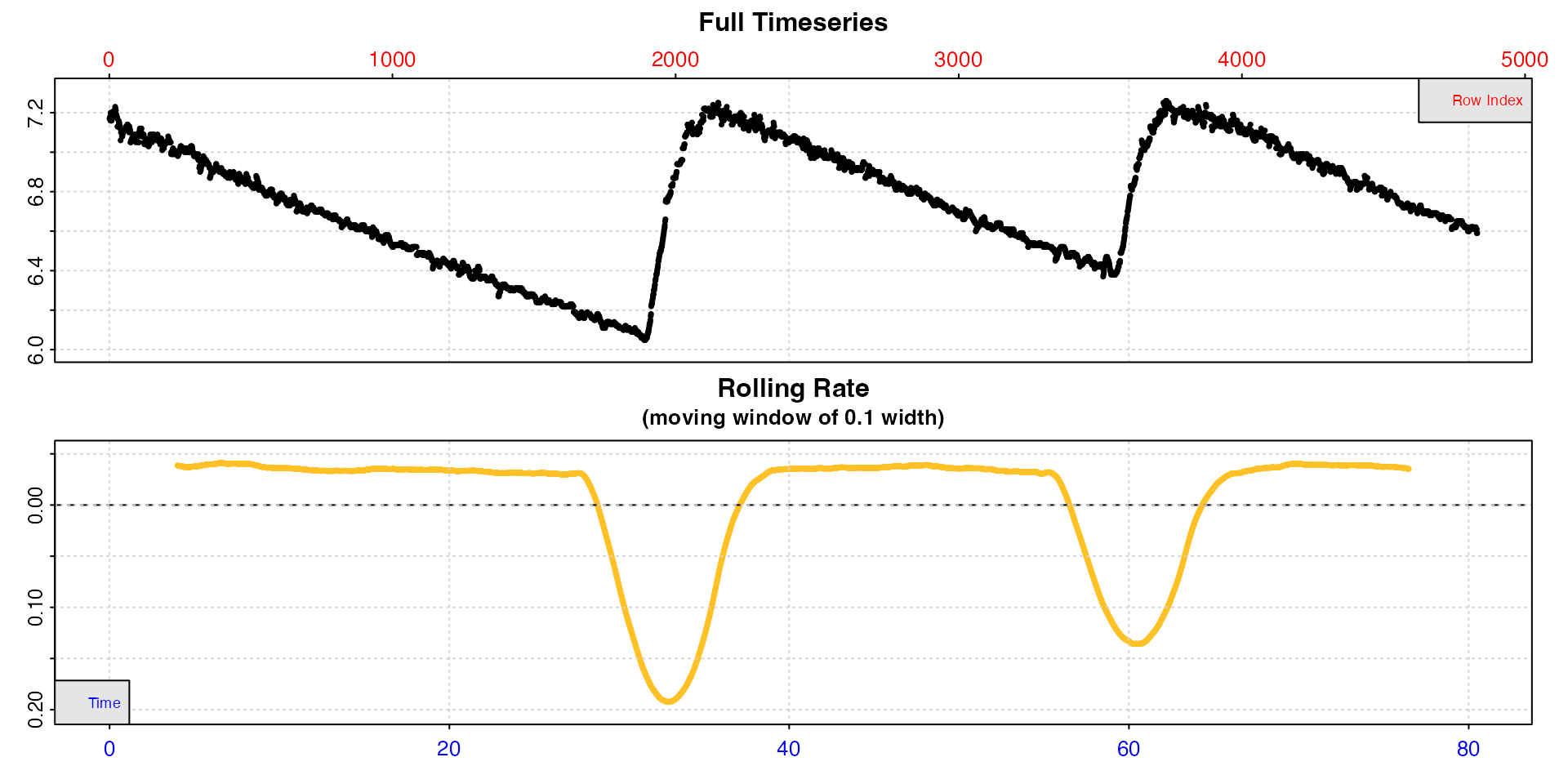
Rate from row range in each replicate
If no inputs other than x and starts (in
the default units of by = "row") are entered, the default
behaviour is to calculate a rate across all the data in each
replicate.
calc_rate.int(urchin,
starts = c(1, 2101, 3901))
#> calc_rate.int: The `measure` input is NULL. Calculating rate to the end of the replicate.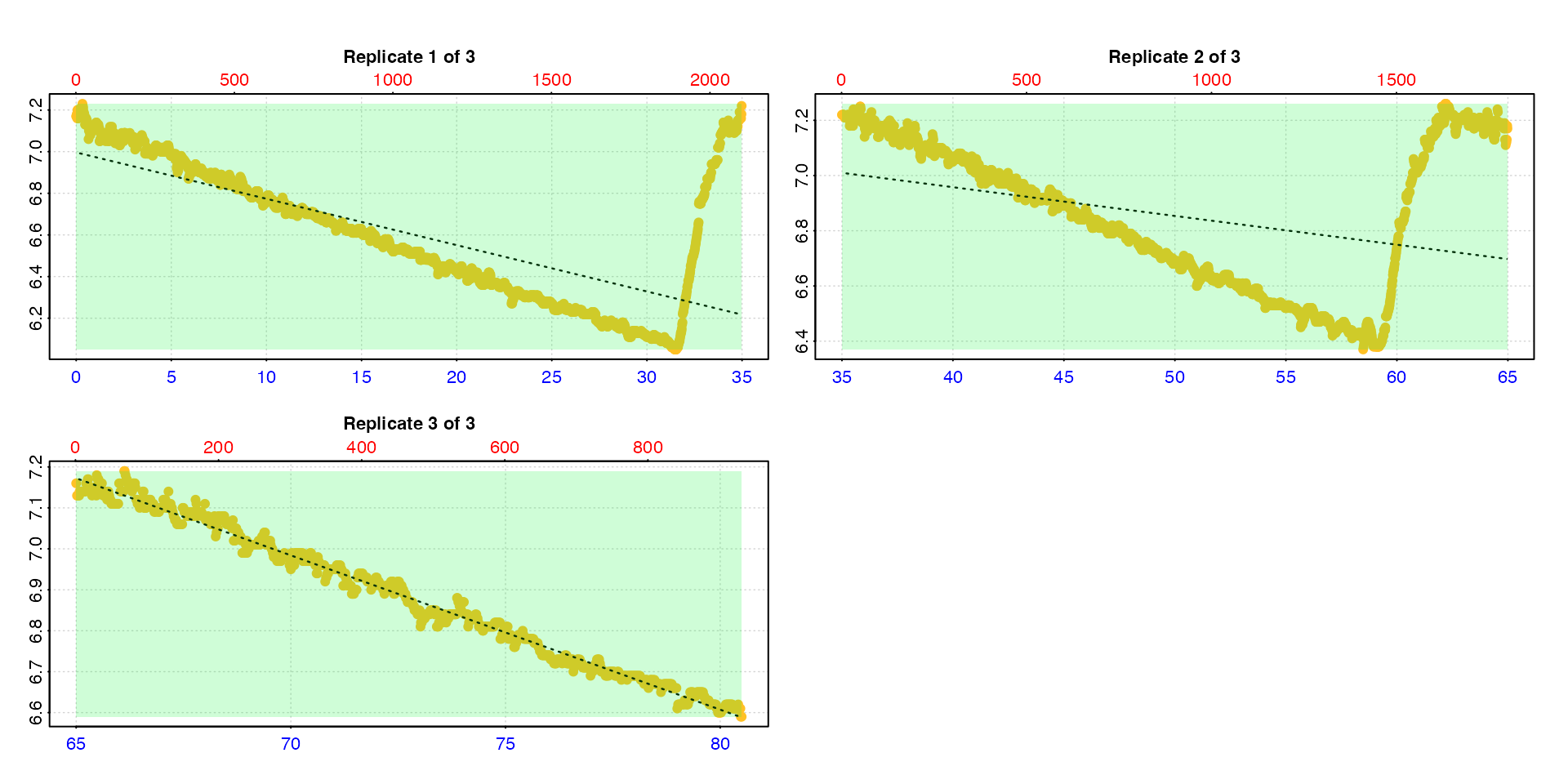
This is obviously not going to produce an appropriate rate. To
exclude flush periods use the measure input. Here, using a
vector of the same length as starts we can specify a
different measure phase in each replicate. The default is
to specify this in row widths, that is by = "row".
calc_rate.int(urchin,
starts = c(1, 2101, 3901),
measure = c(1800, 1200, 800)) 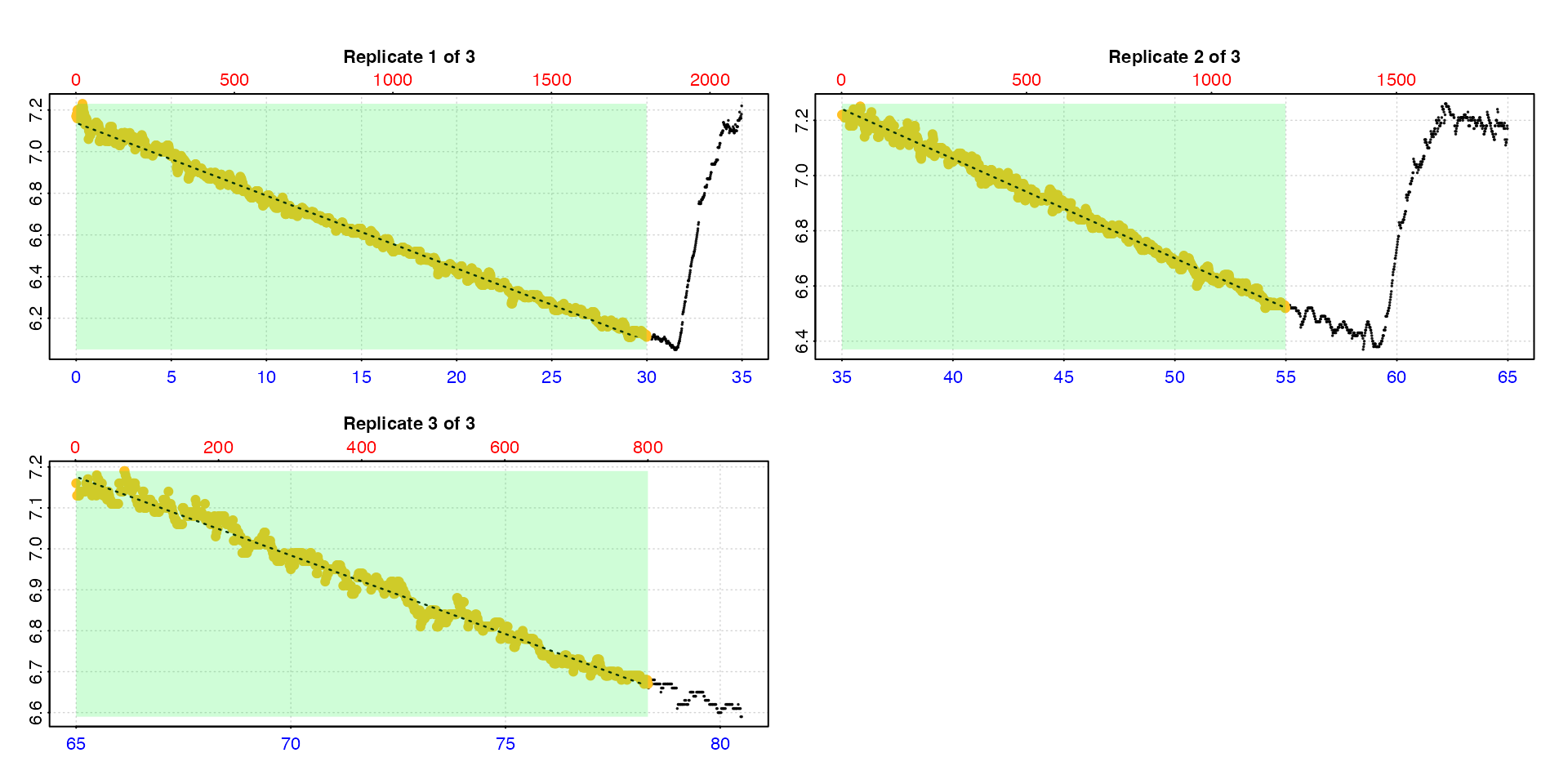
However, we usually want to use the same region within each replicate
to get a rate, and also exclude the first few minutes to allow a period
of settling or acclimation after the flush. We can enter the
wait and measure inputs as single values to
acheive this.
calc_rate.int(urchin,
starts = c(1, 2101, 3901),
wait = 180,
measure = 600) 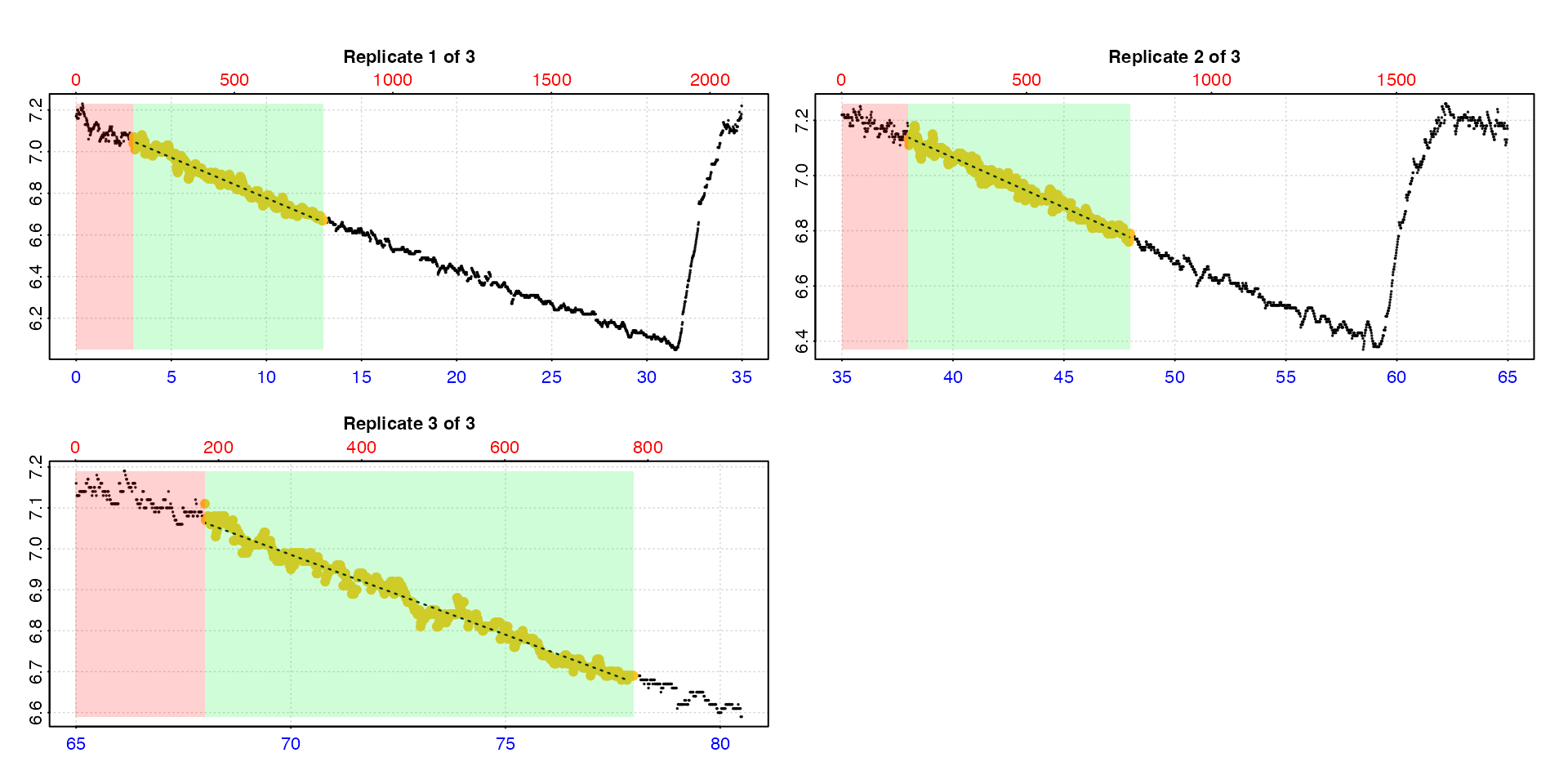
Rate from time range in each replicate
starts, wait, and measure can
also be specified in the time units of the original data by using
by = "time".
calc_rate.int(urchin,
starts = c(0,35,64),
wait = 3,
measure = 10,
by = "time")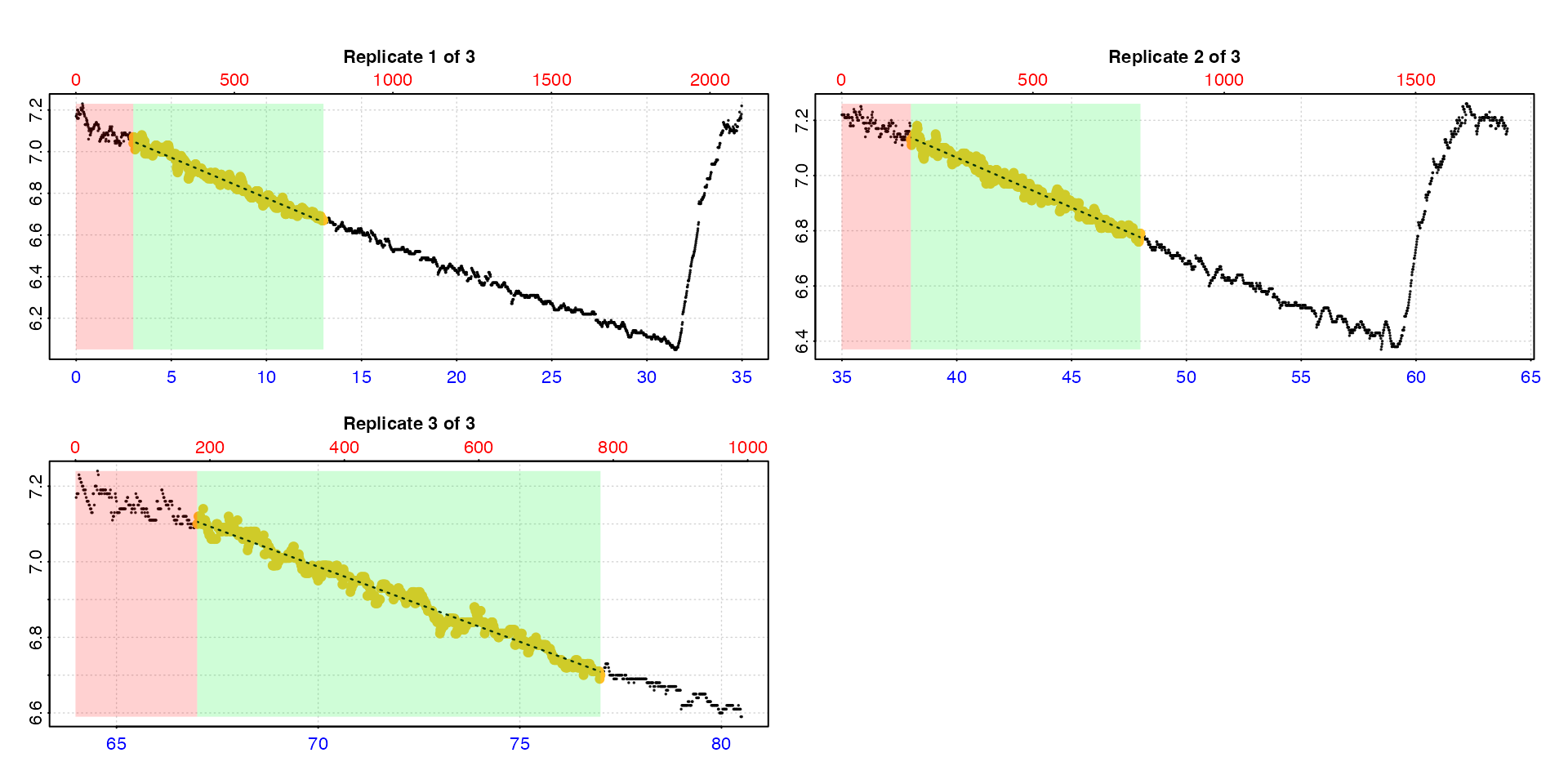
The function uses the closest matching values if the exact values do not occur in the time data.
Regularly spaced replicates
For intermittent-flow experiments where the replicates are regularly
spaced you do not have to specify the start location of every single one
in starts. You can instead enter a value specifying the
interval between each replicate starting at the first row of the input
data in either rows or time units, as specified via the by
input.
Specify regular replicate structure
The zeb_intermittent.rd data is from an
intermittent-flow experiment on a zebrafish. It has a regular replicate
cycle of 11 minutes (660 rows) for most of its length, comprising 9
minutes of measuring and 2 minutes flush. See
help("zeb_intermittent.rd") for exact locations, but
essentially after a background control and a single replicate of 14
minutes this cycle is maintained until the background recording at the
end.
Here we use subset_data() to extract only the regularly
spaced replicates, and pipe the result to calc_rate.int. We
specify the 660 row cycle using starts, exclude the first 2
minutes of each replicate using wait, and extract a rate
from the following 6 minutes using measure, leaving the
flush excluded.
zeb_all <- zeb_intermittent.rd |>
# inspect the data
inspect() |>
# subset regular replicates from larger dataset
subset_data(from = 5840,
to = 75139,
by = "row",
quiet = TRUE) |>
# calc rate in each one from row 120 to 480, plotting first 3 and last 3
calc_rate.int(
starts = 660,
wait = 120,
measure = 360,
by = "row",
plot = TRUE,
pos = c(1:3, 103:105))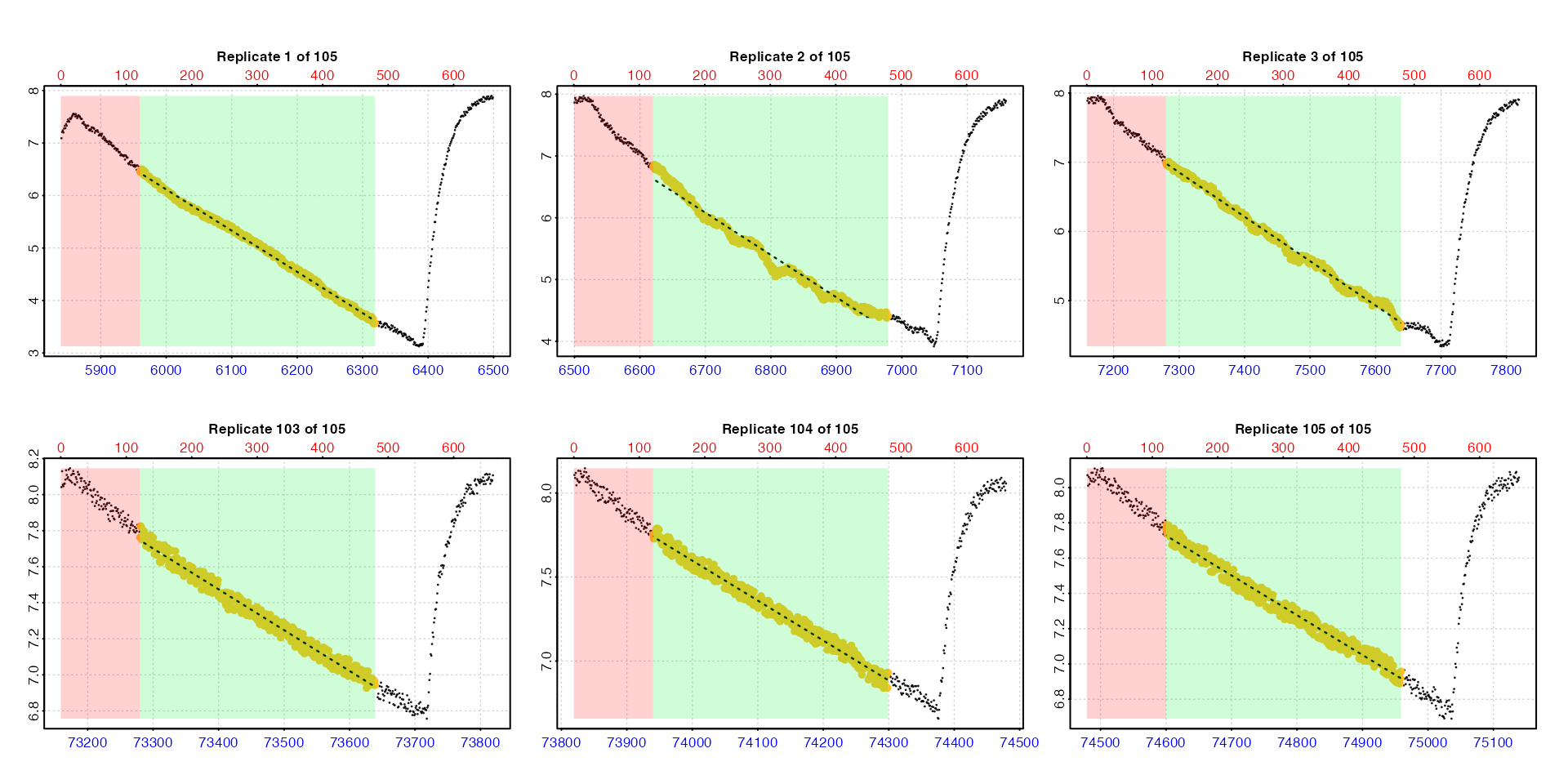
We use the pos input which is passed to
plot to select the first and last three replicates for
plotting to check everything looks okay. In a full analysis however we
strongly recommend every replicate rate result be
examined visually. Up to 20 results at a time can be plotted using the
pos input. See Plot section below for
further details.
View summary
Each replicate result is saved in the $summary element
of the output, or we can use summary().
summary(zeb_all)
#>
#> # summary.calc_rate.int # ---------------
#> Summary of all replicate results:
#>
#> rep rank intercept_b0 slope_b1 rsq row endrow time endtime oxy endoxy rate.2pt rate
#> 1: 1 1 53.2 -0.00785 0.997 121 480 5960 6319 6.45 3.57 -0.00804 -0.00785
#> 2: 2 1 51.9 -0.00684 0.975 781 1140 6620 6979 6.82 4.41 -0.00672 -0.00684
#> 3: 3 1 53.6 -0.00641 0.994 1441 1800 7280 7639 6.99 4.64 -0.00654 -0.00641
#> 4: 4 1 42.1 -0.00442 0.989 2101 2460 7940 8299 7.16 5.45 -0.00478 -0.00442
#> 5: 5 1 40.0 -0.00382 0.989 2761 3120 8600 8959 7.31 5.88 -0.00397 -0.00382
#> ---
#> 101: 101 1 169.3 -0.00225 0.982 66121 66480 71960 72319 7.81 6.94 -0.00241 -0.00225
#> 102: 102 1 178.1 -0.00235 0.984 66781 67140 72620 72979 7.77 6.88 -0.00250 -0.00235
#> 103: 103 1 173.6 -0.00226 0.982 67441 67800 73280 73639 7.76 6.94 -0.00229 -0.00226
#> 104: 104 1 183.5 -0.00238 0.984 68101 68460 73940 74299 7.75 6.93 -0.00229 -0.00238
#> 105: 105 1 175.9 -0.00225 0.980 68761 69120 74600 74959 7.76 6.96 -0.00224 -0.00225
#> -----------------------------------------The replicate number is indicated by the $rep column.
The $rank column indicates ranking or ordering of rates
within each individual replicate. Since
calc_rate.int only returns one rate per replicate it is not
relevant here, but is useful for auto_rate.int() results
where multiple rates per replicate may be returned.
Plot
If plot = TRUE, a plot is produced of each replicate
rate result on a grid up to a total of 20. By default this is the first
20 (i.e. pos = 1:20). Others can be selected by modifying
the pos input, either in the main function call or when
calling plot() on output objects.
plot(zeb_all, pos = 50:69)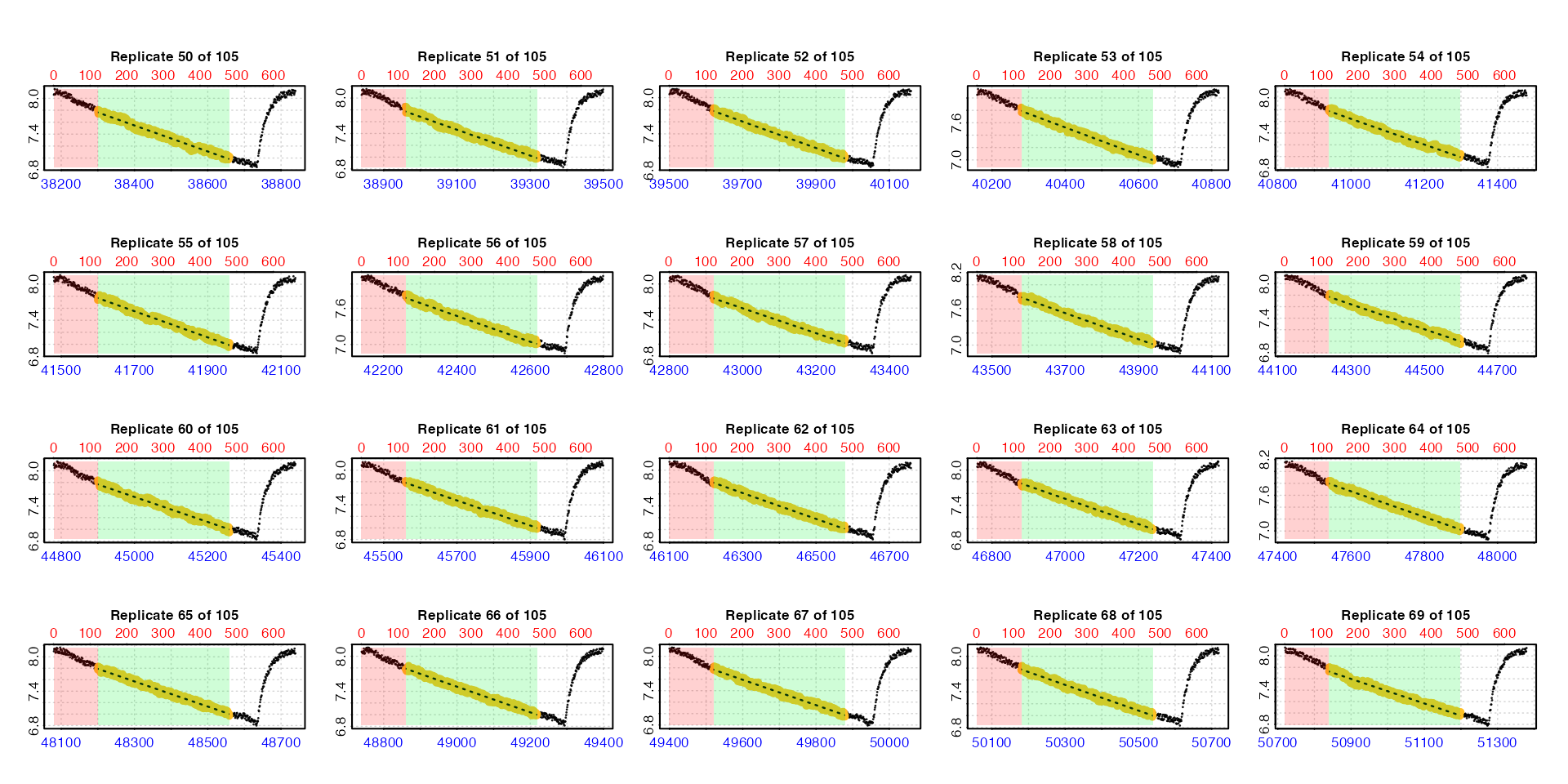
For all plots, the bottom blue time axis shows the time values of the original larger dataset, whilst the upper red row axis shows the rows of the replicate subset.
There are three ways in which these calc_rate.int
results can be plotted, selected using the type input in
either the main function call or when calling plot() on
output objects.
type = "rep"
The default is type = "rep" in which each replicate is
plotted individually with the rate region highlighted.
calc_rate.int(urchin,
starts = c(0,35,64),
wait = 5,
measure = 15,
by = "time")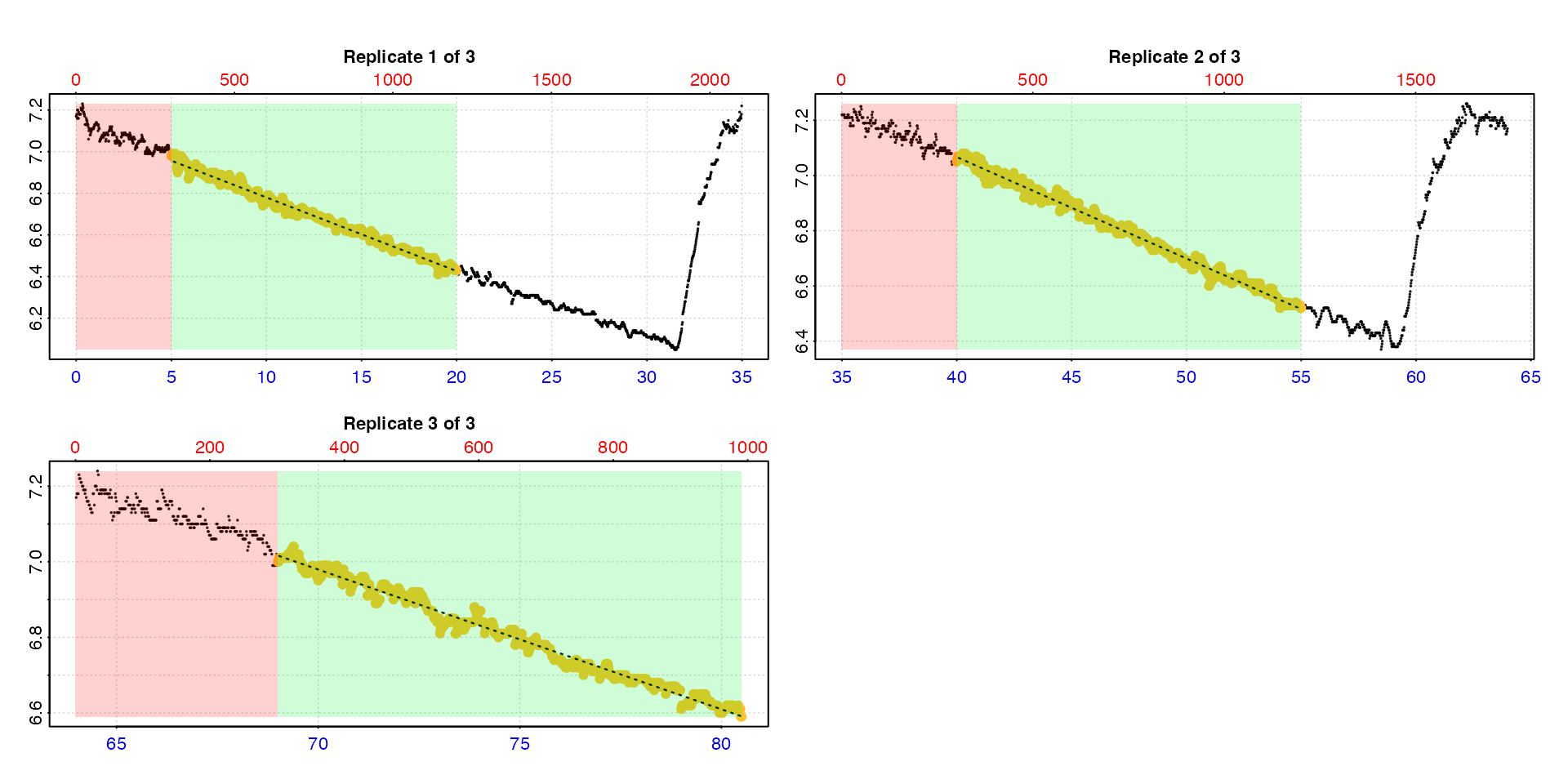
type = "full"
Entering type = "full" will show each replicate rate
highlighted in the context of the entire dataset.
calc_rate.int(urchin,
starts = c(0,35,64),
wait = 5,
measure = 15,
by = "time",
type = "full")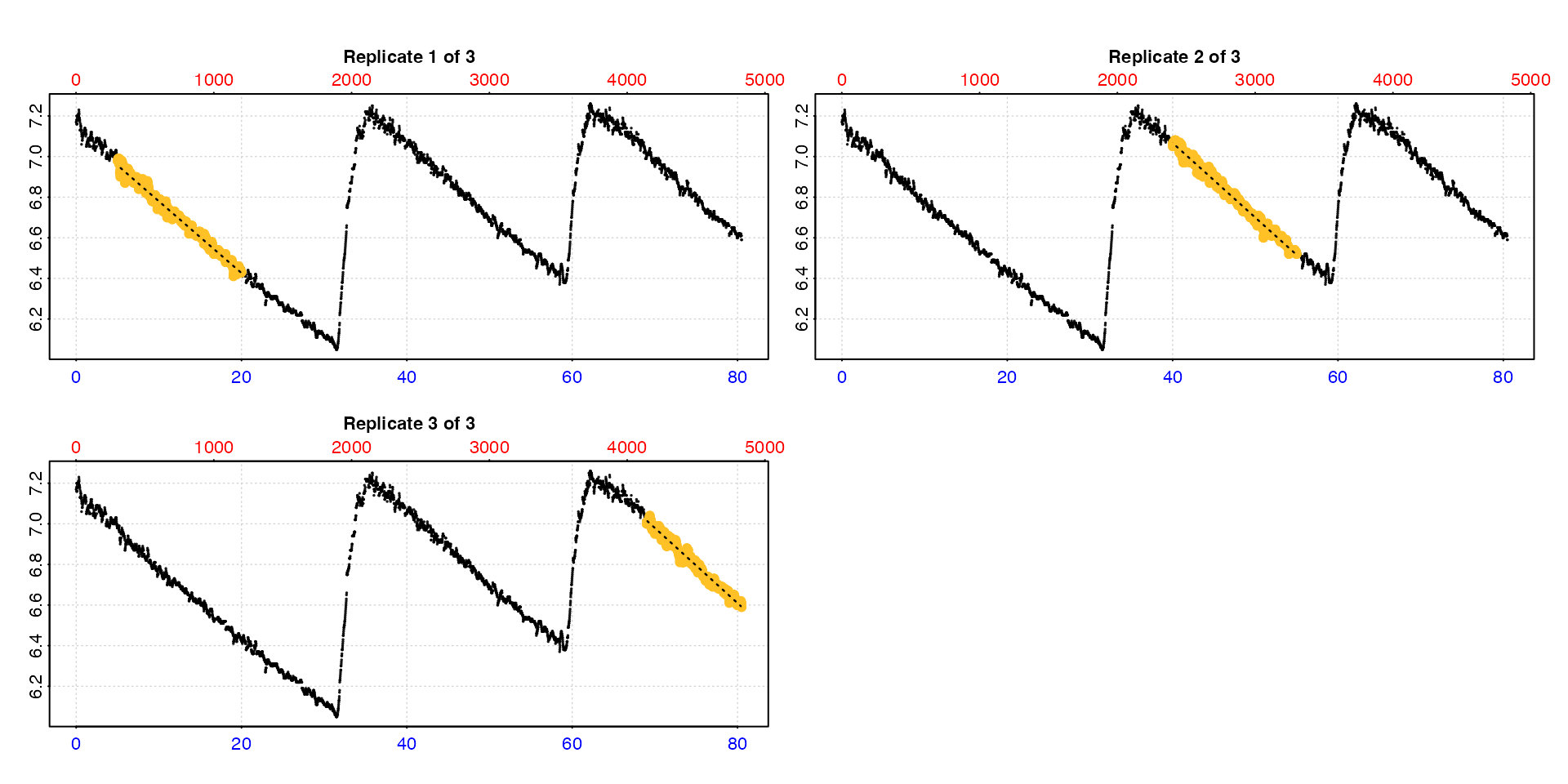
Note this may be of limited use when the dataset is large.
plot(zeb_all, type = "full", pos = 50)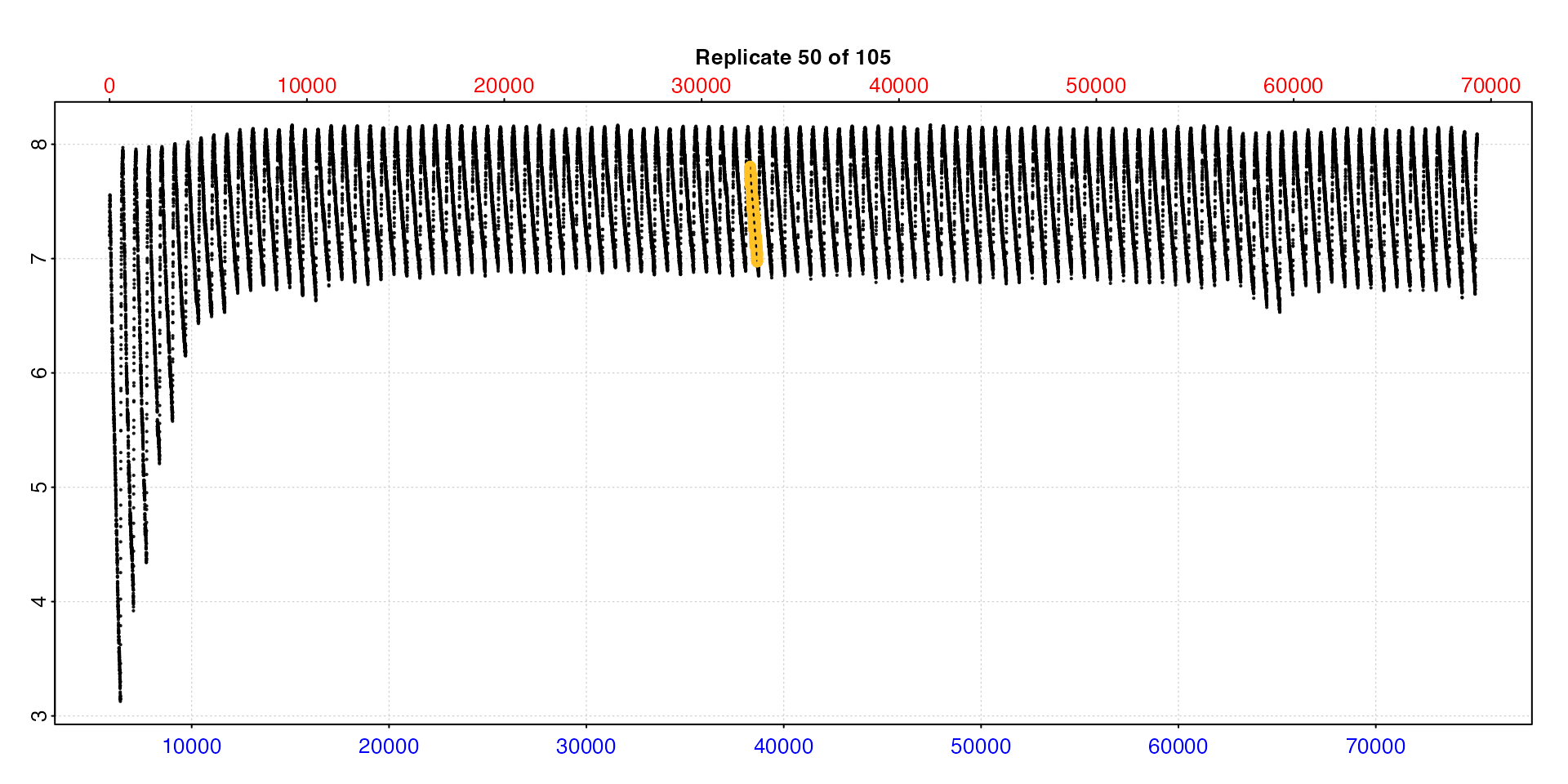
type = "cr"
Lastly, the calc_rate result for each individual
replicate (as found in the $results element of the output)
can be plotted using type = "cr" and the pos
input. Note these are results from subsets of the initial data.
plot(zeb_all, type = "cr", pos = 50)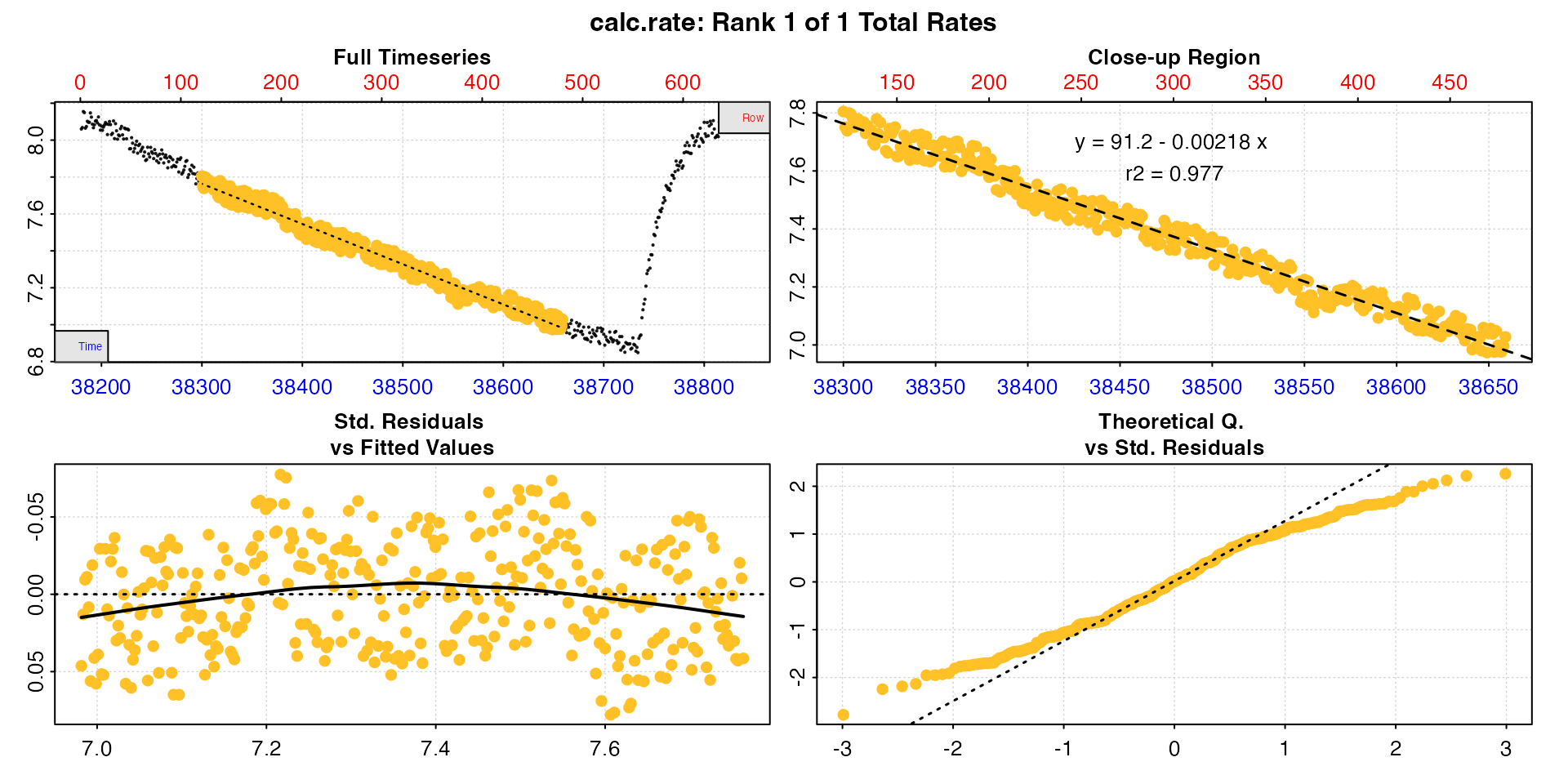
The pos input here can also be of multiple replicates
but this will produce multiple individual plots that you will need to
scroll through.
Additional plotting options
legend can be used to show axis labels (default is
FALSE). quiet suppresses console output.
Additional plotting parameters can also be passed to adjust margins,
axis label rotation, etc. See here.
S3 generic methods
As well as plot, saved calc_rate.int
objects work with the generic S3 methods print,
summary, and mean
print
This simply prints a single replicate rate result to the console, by
default the first one. The pos input can be used to print
others.
print(zeb_all)
#>
#> # print.calc_rate.int # -----------------
#> Replicate 1 of 105 :
#> Rate: -0.00785
#>
#> To see other replicate results use 'pos' input.
#> To see full results use summary().
#> -----------------------------------------
print(zeb_all, pos = 50)
#>
#> # print.calc_rate.int # -----------------
#> Replicate 50 of 105 :
#> Rate: -0.00218
#>
#> To see other replicate results use 'pos' input.
#> To see full results use summary().
#> -----------------------------------------
summary
This prints the summary table to the console which contains linear
model coefficients and other metadata for each replicate rate. The
pos input can be used to select which replicates
($rep column) to include.
summary(zeb_all)
#>
#> # summary.calc_rate.int # ---------------
#> Summary of all replicate results:
#>
#> rep rank intercept_b0 slope_b1 rsq row endrow time endtime oxy endoxy rate.2pt rate
#> 1: 1 1 53.2 -0.00785 0.997 121 480 5960 6319 6.45 3.57 -0.00804 -0.00785
#> 2: 2 1 51.9 -0.00684 0.975 781 1140 6620 6979 6.82 4.41 -0.00672 -0.00684
#> 3: 3 1 53.6 -0.00641 0.994 1441 1800 7280 7639 6.99 4.64 -0.00654 -0.00641
#> 4: 4 1 42.1 -0.00442 0.989 2101 2460 7940 8299 7.16 5.45 -0.00478 -0.00442
#> 5: 5 1 40.0 -0.00382 0.989 2761 3120 8600 8959 7.31 5.88 -0.00397 -0.00382
#> ---
#> 101: 101 1 169.3 -0.00225 0.982 66121 66480 71960 72319 7.81 6.94 -0.00241 -0.00225
#> 102: 102 1 178.1 -0.00235 0.984 66781 67140 72620 72979 7.77 6.88 -0.00250 -0.00235
#> 103: 103 1 173.6 -0.00226 0.982 67441 67800 73280 73639 7.76 6.94 -0.00229 -0.00226
#> 104: 104 1 183.5 -0.00238 0.984 68101 68460 73940 74299 7.75 6.93 -0.00229 -0.00238
#> 105: 105 1 175.9 -0.00225 0.980 68761 69120 74600 74959 7.76 6.96 -0.00224 -0.00225
#> -----------------------------------------
summary(zeb_all, pos = 1:4)
#>
#> # summary.calc_rate.int # ---------------
#> Summary of results from entered 'pos' replicate(s):
#>
#> rep rank intercept_b0 slope_b1 rsq row endrow time endtime oxy endoxy rate.2pt rate
#> 1: 1 1 53.2 -0.00785 0.997 121 480 5960 6319 6.45 3.57 -0.00804 -0.00785
#> 2: 2 1 51.9 -0.00684 0.975 781 1140 6620 6979 6.82 4.41 -0.00672 -0.00684
#> 3: 3 1 53.6 -0.00641 0.994 1441 1800 7280 7639 6.99 4.64 -0.00654 -0.00641
#> 4: 4 1 42.1 -0.00442 0.989 2101 2460 7940 8299 7.16 5.45 -0.00478 -0.00442
#> -----------------------------------------export = TRUE can be used to export the summary table as
a data frame, or those rows selected using pos
zeb_exp <- summary(zeb_all,
pos = 1:4,
export = TRUE)
zeb_exp
#> rep rank intercept_b0 slope_b1 rsq row endrow time endtime oxy endoxy rate.2pt rate
#> <int> <int> <num> <num> <num> <num> <num> <num> <num> <num> <num> <num> <num>
#> 1: 1 1 53.2 -0.00785 0.997 121 480 5960 6319 6.45 3.57 -0.00804 -0.00785
#> 2: 2 1 51.9 -0.00684 0.975 781 1140 6620 6979 6.82 4.41 -0.00672 -0.00684
#> 3: 3 1 53.6 -0.00641 0.994 1441 1800 7280 7639 6.99 4.64 -0.00654 -0.00641
#> 4: 4 1 42.1 -0.00442 0.989 2101 2460 7940 8299 7.16 5.45 -0.00478 -0.00442
mean
This averages all replicate rates in the $rate column,
or those selected using pos. The result can be saved as a
value by using export = TRUE.
zeb_mean <- mean(zeb_all, pos = 1:4, export = TRUE)
#>
#> # mean.calc_rate.int # ------------------
#> Mean of rate results from entered 'pos' replicates:
#>
#> Mean of 4 replicate rates:
#> [1] -0.00638
#> -----------------------------------------
zeb_mean
#> [1] -0.00638This could be passed as a numeric value to later functions such as
convert_rate(), although it is usually better to do this
kind of result filtering after conversion.
Notes and tips
Internally
calc_rate.intusescalc_rate()to extract rates, which explains why each replicate result in$resultsis acalc_rateobject. Thewaitandmeasureinputs are simply converted to thefromandtoinputs ofcalc_rate. Essentiallyfromis equivalent towait, andtoequivalent towait + measure. If you want to use theby = "oxygen"method ofcalc_rateyou will have to look into looping the function. See here.Note the default behaviour of
calc_rateif values are outside the range present within each replicate is used. For example if the enteredmeasurephase is longer than the actual replicate, the last row or time value in the replicate will be used instead. Similarly, ifwaitormeasureinputs do not match exactly to a value within the data the closest matching value is used instead. Seecalc_rate()for full details.The examples above assume flushes occur at the end of each replicate. If your data is subset or processed so that flushes are at the start instead this is easy to deal with. Simply use
waitto exclude the flush at the start plus whatever subsequent wait phase you want to use before themeasurephase. See example here.